Our research is motivated by the dream of identifying novel therapeutic targets for the treatment of acute organ failure in critically ill patients. The most common reasons for admission to the ICU are Sepsis and Acute Respiratory Distress Syndrome (ARDS). Both syndromes affect people of all ages, all ethnicities with no preference for gender or pre-clinical status. Despite enormous efforts, expertise and resources about 40% of patients die, and those who survive experience significant long term morbidity including muscle weakness, cognitive impairment and a persistently elevated risk of death. Success also comes at a significant societal cost: ICU care alone consumes 8% of the hospital budget and in Canada about 0.2% of our GDP, or $4 billion annually. On a global level, these costs are staggering rising to hundreds of billions of dollars annually. More importantly, the burden to patients, their loved ones and care givers remains a challenge. Most patients who die with sepsis or ARDS, die from multi-organ dysfunction syndrome (MODS). MODS is a continuum, with incremental degrees of physiologic derangements in individual organs; it is a process rather than a single event. Not all organs fail equally, and not all processes of failure are similar. Alteration in organ function can vary widely from a mild degree of organ dysfunction to completely irreversible organ failure; but the degree of organ dysfunction has a major clinical impact and understanding how this happens will provide novel insights regarding how to prevent, treat and reverse organ injury in these patients.
My laboratory is dedicated to identifying putative genes/molecules, pathways and networks involved in the pathophysiology, diagnosis, prognosis and management of critical illness-induced acute organ failure: specifically lung, muscle and heart.
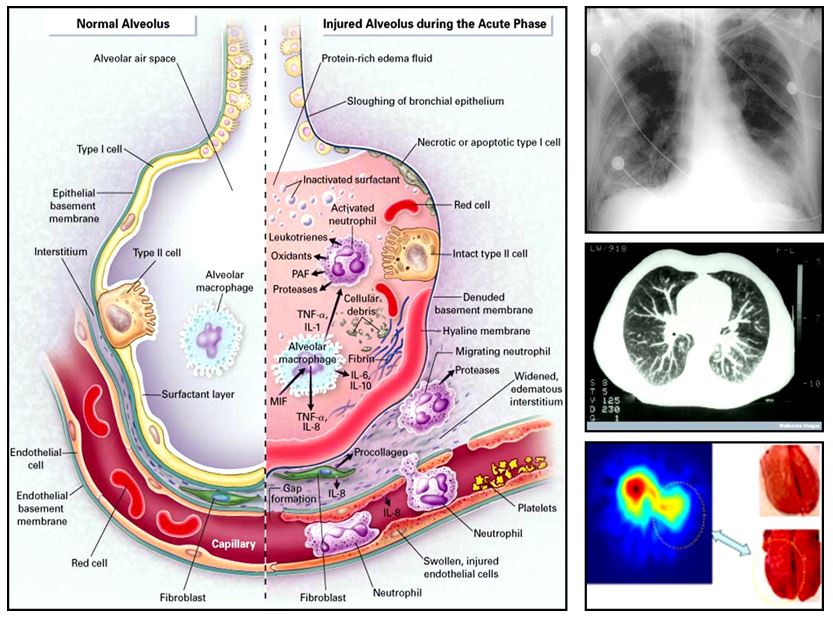
Acute Lung Injury (ALI): The response to injury is a complex, multigenic event, involving multiple factors (dos Santos et al. CCM 2008), but what seems to be clear is that the mechanical ventilator exerts an important immunomodulatory function. Cyclical overdistension and/or cellular deformation have been shown to augment the inflammatory response in critically ill patients. The coordinated response of lung cells to cyclic stretch is regulated by the stretch sensitive transcription factor activating transcription factor 3 (ATF3, Akram et al. AJRCCM 2010). Chimeric mice generated by adoptive bone marrow transfer demonstrated that while ATF3 in myeloid-derived cells functions as a negative transcriptional regulator of inflammation, resident cells rely on ATF3 to mount a “defense” response in an Nrf2 dependent manner (Shan Y et al. ARS 2015). Microarray gene-gene meta-analysis shows the potential to differentiate clinically relevant ALI using expression profiles (P. Hu et al. Plos One, 2012).
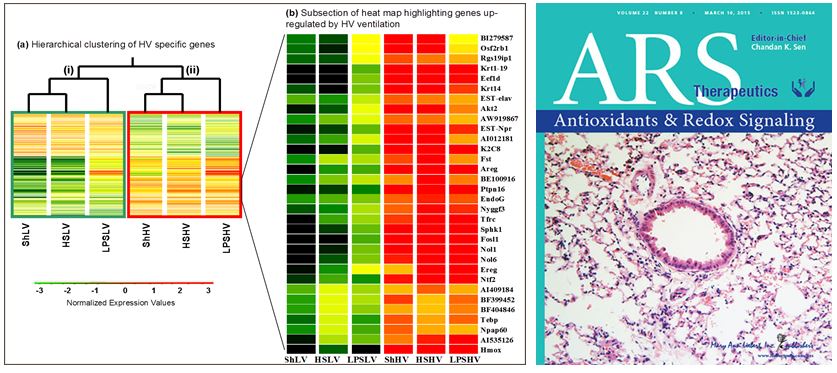
Mesenchymal Stromal/Stem Cells (MSCs) in Multiorgan Injury: Recently, MSCs have been shown to modulate the inflammatory and innate response to injury, sepsis and acute lung injury. In collaboration we have shown that MSCs significantly decreased mortality in septic mice. Analysis of expression profiles from treated vs. untreated mice demonstrated downregulation of inflammation-related genes, and a shift towards upregulation of genes involved in bacterial killing indicating beneficial effects of MSCs extend beyond suppression of inflammation (Mei et al. AJRCCM 2010). Global expression analysis points to a fundamental role of mitochondria related genes (dos Santos et al. Am J of Pathol, 2012). Gene product of the Parkinson disease (autosomal recessive, early onset) 7 [PARK7] or DJ-1 plays a fundamental role in bacterial clearance and survival from sepsis (Amatullah et al. AJRCCM, 2017). MSCs can also attenuate fibroproliferative changes in the lung that occur secondary to dysregulated repair of lung injury (Maron-Gutierrez et al. Crit Care Med. 2013). MSCs can be genetically modified to enhance their therapeutic potential (Zuckermann et al. in preparation) and RNA-based therapeutics may further enhance beneficial effects of MSCs in vivo (Barker et al. in preparation). Importantly, our first publication as collaborators on the Cellular Immonotherapy for Sever Sepsis (CISS) trial, has been submitted for publication. This study performed under the auspices of the Canadian Critical Care Translational Biology Group is the first in-human study exploring the application of mesenchymal Stromal/Stem Cells for the treatment of severe sepsis.
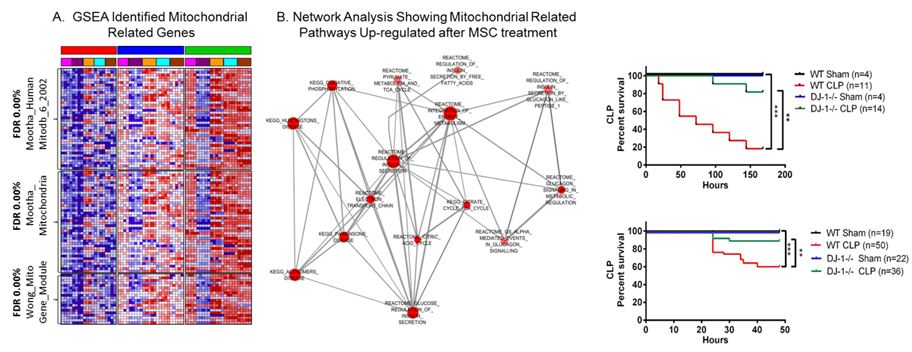
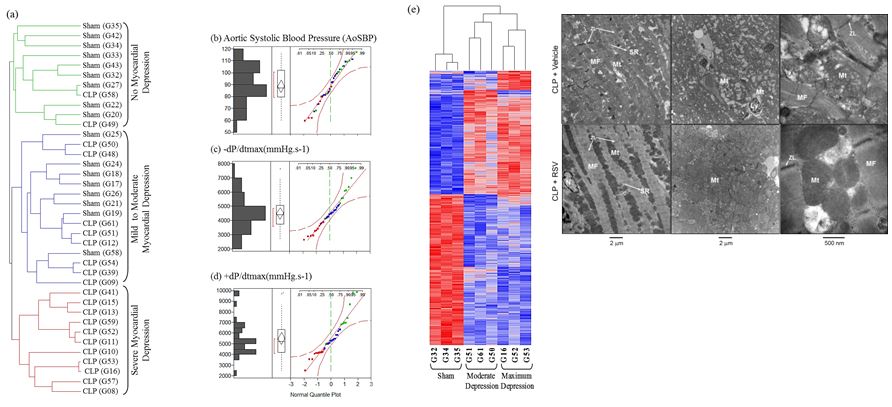
Sepsis-induced Myocardial Depression: Sepsis-induced myocardial depression results in down regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1a). Mitochondrial resuscitation with Resveratrol – antioxidant that increases PGC1a – protects from myocardial depression, suggesting preservation of PGC1a represents a potential treatment strategy for myocardial depression (Smeding L et al. CCM 2012). Targeted deletion of DJ-1 abrogates sepsis-induced myocardial depression and prevents vasoplegia following experimental sepsis (Amatullah et al. in preparation).
ICU Acquired Weakness: In collaboration with Drs. Jane Batt and Margaret Herridge (Batt et al. AJRCCM 2013; Batt et al. JAMA 2013; dos Santos et al. Curr Opin Crit Care 2012) we performed a nested prospective pilot study of patients recruited from Towards RECOVER (Herridge et al. AJRCCM 2016) involving comprehensive assessment of muscle function and molecular determinations (dos Santos et al. 2016). Microarray analysis of gene expression profiles from biopsy samples collected from either healthy controls, 7 days and 6 months ICUAW patients demonstrated clustering of transcriptional profiles according to muscle mass and function. Functional enrichment shows down regulation of both mitochondrial and biosynthetic pathways compared to healthy controls suggesting abnormal cellular energetics and synthetic status (Walsh et al. Scientific Report, 2016).
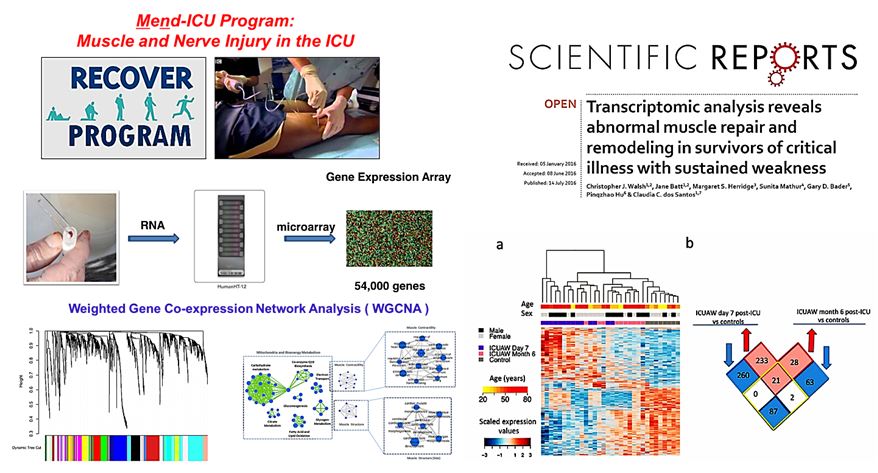
Bioinformatics and Computational Strategies Application: We focused on gene set and regulatory element analysis to identify critical “hub” genes/pathways, gene-sets and regulatory element involved in phenotype determination (Walsh CJ et al, Cancer Informatics 2016, Walsh CJ, et al. Scientific Report, 2016, and Khan A. et al. PlosOne. 2013). Combining this approach with gene-gene mata-analysis to capitalize on all the electronic data currently available (Hu et al. PlosOne. 2013) and develop and understanding of network analysis of transcriptional responses (dos Santos et al. Am J Pathol, 2012, dos Santos CC, Liu M. Mathematical and Computational Biology, 2007). In addition, we have focused on gene ‘interaction’ networks rather than transcriptional changes to identify novel putative targets for therapy (Abdala et al, in preparation).