There are many applications for polymerase chain reaction (PCR) in molecular research such as genotyping, mutagenesis, DNA and RNA amplification and introducing restriction sites to cDNA for sub cloning, to name just a few.
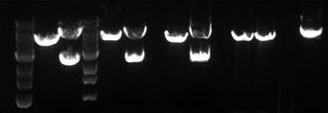
Fig. 1 – Agarose gel of various PCR products
Most PCR reactions are performed with similar reagents.
| 10X PCR Buffer | 1X |
| 10 mM dNTP mixture | 0.2 mM each |
| 50 mM MgCl2 | 1.5 mM |
| Primer mix (5 µM each) | 0.33 µM each |
| Template DNA | ~50 ng |
| DNA Polymerase | Units depend on application |
| Autoclaved, distilled water | Not applicable |
… and they follow the same basic thermocycling protocol of (25-40) repeating cycles:
The success of any PCR reaction is directly related to the specificity and thermal properties of the primers used in the reaction.
When designing primers, consideration should be given to the area of the target gene being amplified (target sequence) and the thermodynamic properties of the primers themselves. All of these properties factor into the efficiency and specificity of target gene amplification. Forward and reverse primer pairs for PCR should bind to unique target sequences between 18-30 bp long that are areas of low secondary structure (GC content < 60%). The primers themselves should follow the guidelines for good primer design (see below), but briefly, they should have low secondary structure (hairpins, loops etc..) and should be “anchored” with a G/C at the 5’ end of the sequence with their melting temperature approximately 5°C higher than their annealing temperature. Primer pairs should have similar thermodynamic properties. There are many good on-line sites for primer design:

Fig 2. – Sample read-out from “Primer BLAST” primer design program
Aim for 18-30 nucleotides in length
Try to make sure the melting temperatures are between 55° and 65°C and, for a pair, that they are within 5°C of each other
If you have a low Tm primer, find a sequence with more Gs and Cs, or extend the length of the primers slightly
Avoid intra-primer complementary (where there are more than three bases that complement within the primer) or inter-primer complementary (where forward and reverse primers have complementary
Typically, 3-4 nucleotides are added to the 5’ of the restriction site in the primer to allow for efficient cutting
Aim for the GC content to be 40-60%, with a C or G at the 3’ end to promote binding
Avoid regions of secondary structure and have a balanced distribution of GC-rich and AT-rich domains
Try to steer clear of runs of four or more of one base, or dinucleotide repeats
If you are using the primers for cloning, we recommend a minimum level purity by cartridge purification
If you’re using your primers for mutagenesis, have the mismatched bases toward the middle of the primer
**Remember to Blast your primer sequences to confirm recognition of the desired template**