NanoString nCounterTM technology is a direct digital detection system, which enables both highly sensitive and reproducible multiplexed gene quantification without amplification. It measures nucleic acid using fluorescent probes which bind directly to chosen targets: mRNA, miRNA, or DNA. This technology can measure up to 800 genes in a single reaction, which not only greatly reduces the number of reactions required, but saves on the required RNA/DNA input amount.
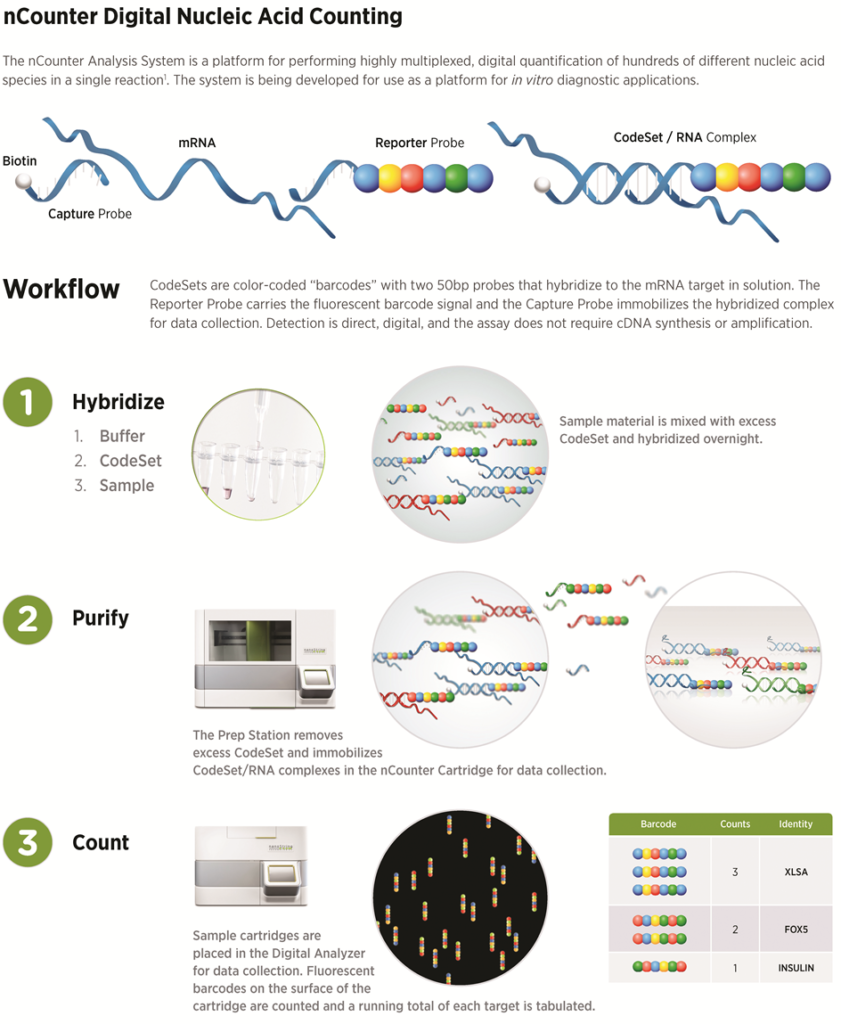
Each gene of interest requires a corresponding capture probe and reporter probe. The reporter and capture probes for all genes of interest are collectively known as the Code Set. The first probe, or capture probe, contains a 35-50 base pair sequence complementary to a target mRNA with a short common sequence coupled to an affinity tag, such as biotin. This capture probe, specifically the affinity tag, allows the complex to be immobilized on the cartridge for data collection. The second probe, or reporter probe, contains a second 35-50 base pair sequence complementary to the target mRNA, which is coupled to a color-coded (fluorescent) tag that provides the detection signal. The probes are hybridized directly to one of several targets (i.e., RNA, miRNA or DNA), where a stable tripartite structure (target/capture/reporter) is formed and linked to a streptavidin-coated cartridge. With the fixed structure, the fluorescent tags can be counted for data analysis.
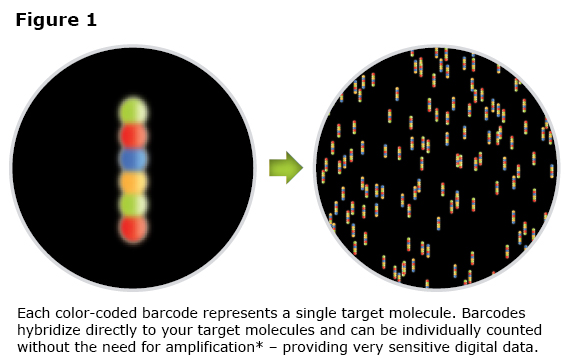
The Digital Analyzer counts the number of fluorescent tags bound. The Digital Analyzer utilizes an epi-fluorescent microscope with a high magnification lens and CCD camera to collect image data (See Figure 1) and then converts it to a digital signal.
The workflow is as follows:

Service includes:
The price of service depends on the scope of the work. Please contact us (pamela.plant@unityhealth.to) for more information.
Reagent pricing: Reagents are purchased directly through Nanostring. Click here for reagent price list or contact info@nanostring.com
Sample requirements:
The prep station will process 12 samples (per cartridge) at any given time. 6 cartridges can be put in the nCounter (counting) station. Pre-fabricated Code Sets can be purchased in quantities of 12, 24, 48, 96, 192 and 384 (and even higher in multiples of 384).
All custom and pre-fabricated nCounter code sets include positive controls and synthetic RNA targets that have been spiked in at a range of concentrations for Quality Control (assessing the success of the run). Negative controls (sequences that should not cross hybridize to the genome/ transcriptome) are also included. Housekeeping genes are included in pre-fabricated code sets that can be used for normalization but these have to be designed into custom sets.
Nanostring offers routine and advanced analysis modules which run in the nSolver software. You can also choose to export the data (digital counts) from an nCounter experiment to a CSV file, which can be imported into third party analysis program (i.e. Gene Pattern, Broad Institute or Matlab etc.). Data analysis manuals are available for all applications on the Nanostring website and are excellent resources for guidelines on normalization and data analysis.