 Next-generation sequencing refers to non-Sanger-based high-throughput DNA sequencing technologies. Millions or billions of DNA strands can be sequenced in parallel, yielding substantially more throughput. Sequencing may be utilized to determine the order of nucleotides in small targeted genomic regions or entire genomes. Illumina sequencing enables a wide variety of applications, allowing researchers to ask virtually any question related to the genome, transcriptome, or epigenome of any organism. Next-generation sequencing (NGS) methods differ primarily by how the DNA or RNA samples are prepared and the data analysis options used.
Next-generation sequencing refers to non-Sanger-based high-throughput DNA sequencing technologies. Millions or billions of DNA strands can be sequenced in parallel, yielding substantially more throughput. Sequencing may be utilized to determine the order of nucleotides in small targeted genomic regions or entire genomes. Illumina sequencing enables a wide variety of applications, allowing researchers to ask virtually any question related to the genome, transcriptome, or epigenome of any organism. Next-generation sequencing (NGS) methods differ primarily by how the DNA or RNA samples are prepared and the data analysis options used.
The workflow of NextSeq can be divided into four major steps:
Samples consisting of longer fragments are first sheared into a random library of 100-300 base-pair long fragments. After fragmentation the ends of the obtained DNA-fragments are repaired and an A-overhang is added at the 3′-end of each strand. Afterwards, adaptors which are necessary for amplification and sequencing are ligated to both ends of the DNA-fragments. These fragments are then size selected and purified. 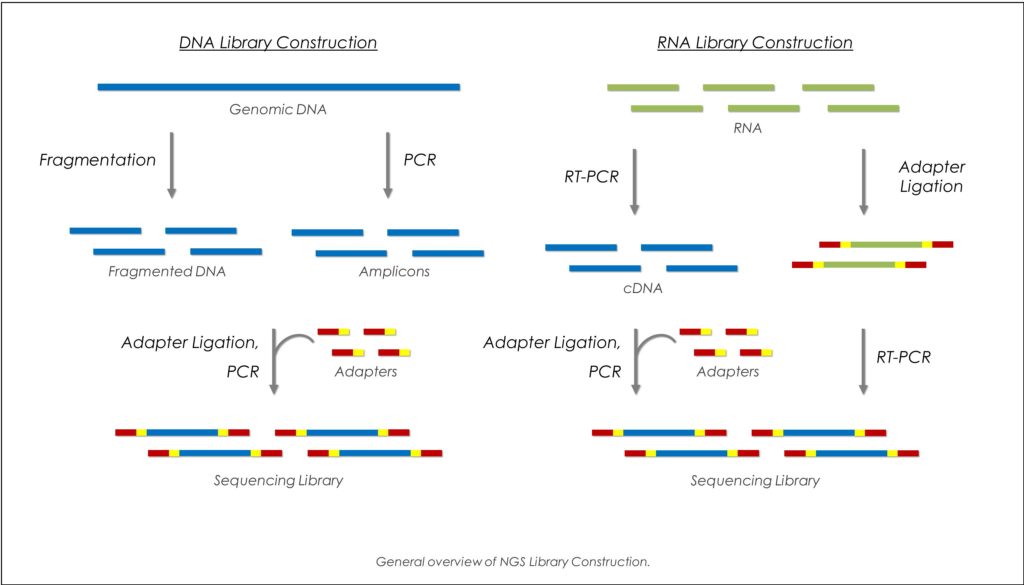
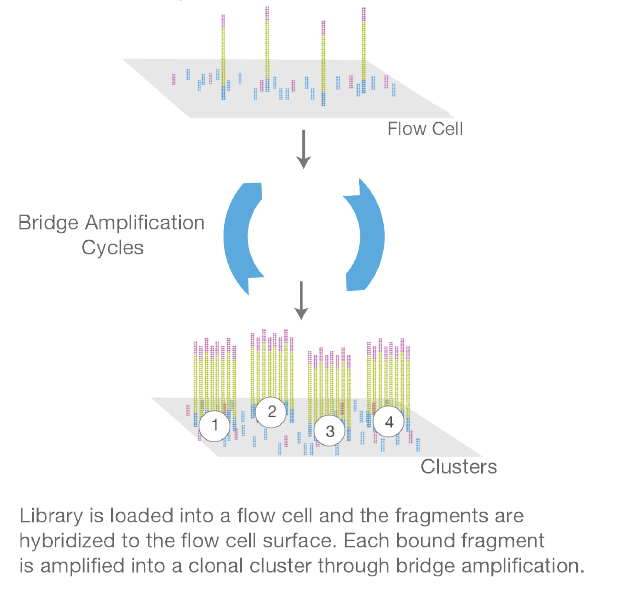
The Cluster Generation is performed on the Illumina NextSeq 550. Single DNA-fragments are attached to the flow cell by hybridizing to oligos on its surface that are complementary to the ligated adaptors. The DNA-molecules are then amplified by a so called bridge amplification which results in a hundred of millions of unique clusters. Finally, the reverse strands are cleaved and washed away and the sequencing primer is hybridized to the DNA-templates.
During sequencing the huge amount of generated clusters are sequenced simultaneously. The DNA-templates are copied base by base using the four nucleotides (ACGT) which are fluorescently-labeled and reversibly terminated. After each synthesis step, the clusters are excited by a laser which causes fluorescence of the last incorporated base. After that, the fluorescence label and the blocking group are removed allowing the addition of the next base. The fluorescence signal after each incorporation step is captured by a built-in camera, producing images of the flow cell. After sequencing, with QC pass and FASTQ data will be generated, integrated data analysis can be applied as your project. 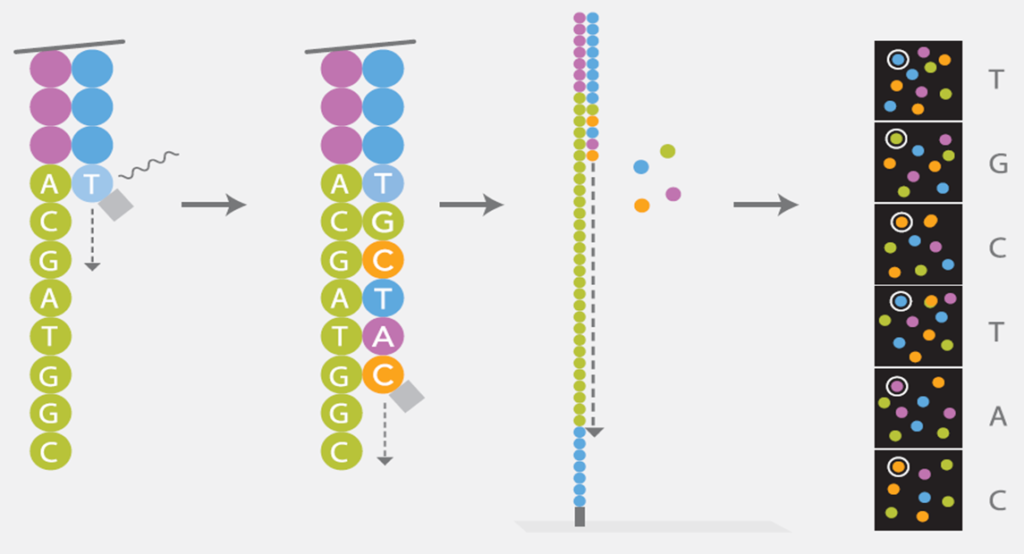
Reads are aligned to a reference sequence with bioinformatics software. After alignment, differences between the reference genome and the newly sequenced reads can be identified. 
mRNA sequencing (mRNA-Seq) enables researchers to take a closer look at gene expression and can be used to identify isoforms, novel transcripts, and gene fusions. The NextSeq RNA sequencing solution leverages proven Illumina technology to deliver a detailed snapshot of the coding transcriptome. Sequence up to 16 transcriptomes per NextSeq run.
mRNA library Prep kit: Prepare sequencing libraries from mRNA to get a clear view of the coding transcriptome with strand-specific information.
NextSeq 550 v2 Reagent kits: Bring the power of a high-throughput sequencing system to the benchtop. The 150 cycle high-output or mid-output kits provide 2 x 75 bp reads for total RNA-Seq.
BaseSpace RNA-Seq Alignment App: Performs read mapping, normalized abundance estimation of reference genes and transcripts, novel transcripts assembly, variant calling, and optional fusion calling. BaseSpace Cufflinks Assembly & DE App: Quickly assess novel transcript isoforms and gene expression levels from RNA-Seq Alignment results.
Uses a simple to provide a fast path to greater understanding of the exome.
NextSeq 550 v2 Reagent kits: Bring the power of a high-throughput sequencing system to the desktop. The 300 cycle high-output or mid-output kits provide 2 x 150 bp reads for exome sequencing.
BaseSpace Enrichment App: Rapid alignment and variant detection for small, structural, and copy number variant calling, variant annotation, and enrichment metrics calculation.
TruSeq Nano DNA Library Prep Kit: Generate WGS libraries and efficiently interrogate samples with limited available DNA. Nextera DNA Flex Library Prep Kit: A fast, integrated workflow for a wide range of applications, from human whole-genome sequencing to amplicons, plasmids, and microbial species.
NextSeq 500/550 v2 Reagent Kits: Bring the power of a high-throughput sequencing system to the desktop. The 300 cycle high-output kit provides 2 x 150 bp reads for whole-genome sequencing.
BaseSpace Whole-Genome Sequencing App: Quickly extracts biological information from whole-genome sequences, using Isaac alignment and variant calling.
Isolate and sequence small RNA species, such as microRNA, to study the role of noncoding RNA in gene silencing and posttranscriptional regulation.
Analyze both coding RNA and multiple forms of noncoding RNA with total RNA sequencing, for a comprehensive view of the transcriptome.
Detect both known and novel features of the RNA exome using sequence-specific capture of RNA coding regions.
Targeted resequencing focuses time, expenses, and analysis on sequencing only a subset of genes or genome regions of research interest.
Targeted gene sequencing panels contain defined probe sets focused on specific genes of interest. Both predesigned and custom panels are available.
Perform epigenetic studies with high-coverage density and flexibility enabled by sequencing-based DNA methylation analysis.
Ultra-deep sequencing of PCR amplicons enables cost-effective analysis of up to hundreds of target genomic regions in one assay.
Genotyping by sequencing provides a low-cost genetic screening method to discover novel plant and animal SNPs and perform genotyping studies.
Next-generation sequencing offers base-by-base, genome-wide detection of chromosomal variation to complement array-based cytogenomic analysis.
The price of service depends on the scope of the work. Please contact us for more information.
Reagent pricing: Reagents are purchased directly through Illumina or other comparable company, like NEB.