In addition to determining DNA content, one can also determine cellular proliferation in terms of the number of times a cell has divided. Often people confuse the two and think they must give the same information, which is incorrect. Cell cycle analysis tells you how many cells are in each phase (G, S, G2/M) which is like looking at a car race and seeing how many cars are in a pit-stop, on the straightaways or slowing down for turns. This does NOT tell you how many laps each car has done (analogous to number of cell divisions a cell has undergone) or who is winning the race. Cell proliferation analysis does just that via labeling a starting cell population with a dye such as CFSE, PKH dyes, etc. As the cell divides, the fluorescence in the daughter cells will be half as bright as the parent cell. In this manner you can see how many times a cell has divided and calculate meaningful quantitative parameters such as the Proliferation Index.
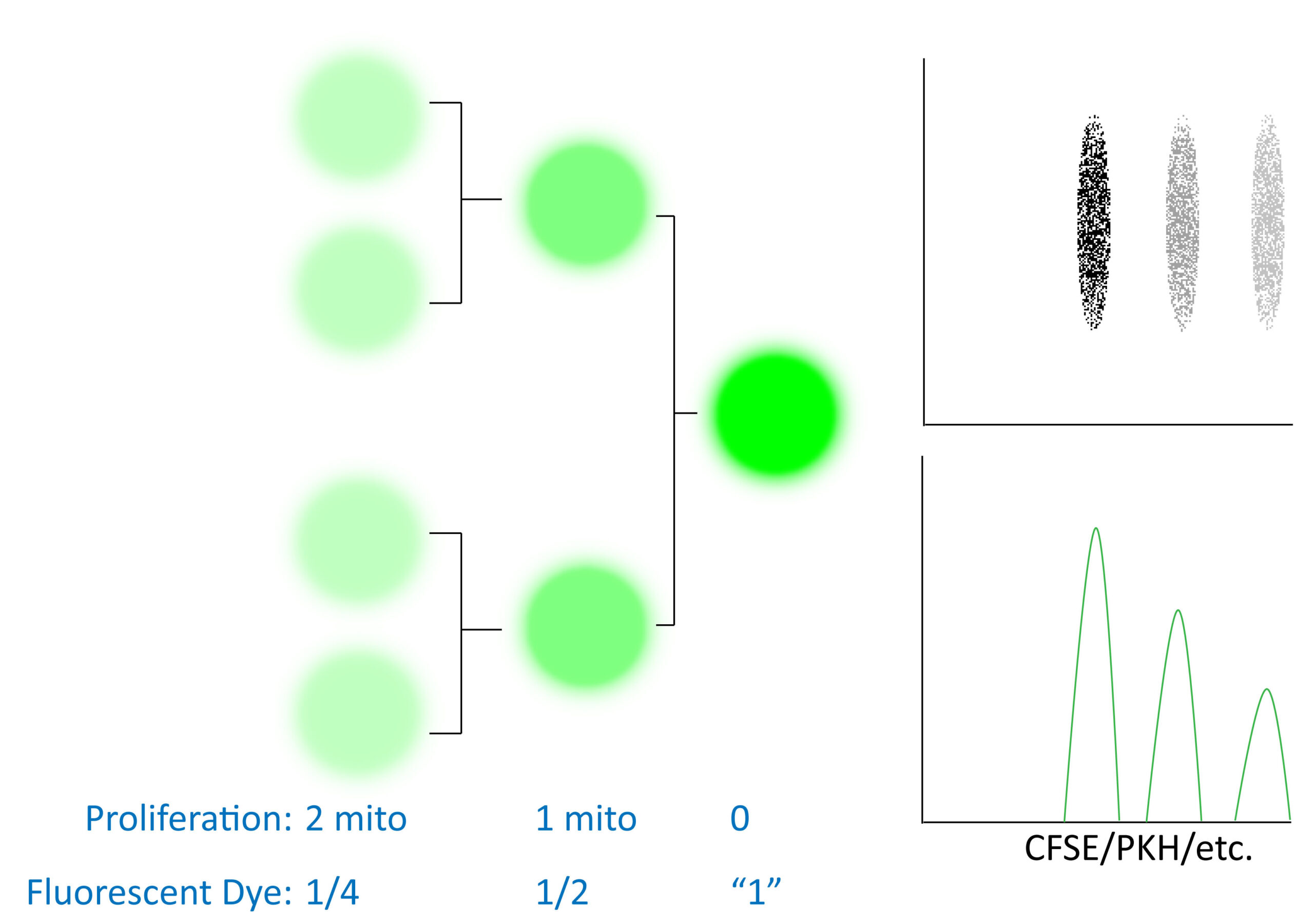
Cellular proliferation dyes are generally quite stably integrated (commonly intracellular protein esterified fluororescent dyes or membrane lipid integrating dyes) and as a result you can track cellular division over several generations. Depending on the proliferation rate of your cell population of interest, it may be possible to track your cells and their proliferation status over long periods of time in vivo. This type of experiment is distinct from MTT assays which are more of an indication of cellular respiration and cell cycle analysis (not how many times a cell has divided, but what portion of cells are in each stage of the cell cycle). Fluorescent dye dilution experiments instead report how many rounds of mitosis a cell has undergone (often as a marker of transformation or cellular activation). At high levels these dyes (CFSE, etc) can be toxic and/or leak out of cells over time. As such, it is recommended to optimize your staining, experimental treatments/timepoints for your cell type of interest before starting a large scale experiment. Some experimental recommendations are also captured in an excellent presentation in the Links section below.
There are several key elements to performing good labeling of cells for proliferation analysis (Figure 2). In particular it is crucial to:
Without taking into consideration these suggestions, it is likely that your proliferation data will not look like publishable data and key metrics like the proliferation index will be very inaccurate (see below).
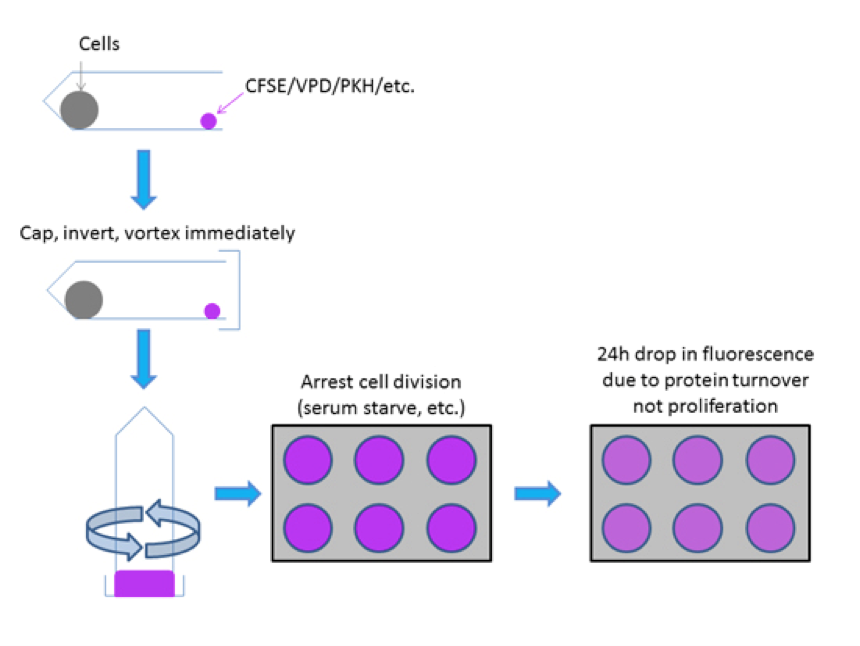
Through rapid mixing of a single cell suspension with CFSE or other proliferation dye (i.e. VPD450 etc) even labeling can be achieved to resolve distinct generations of undivided and divided cell populations which are necessary to empirically measure cell proliferation. It is critical to control for the first 24h drop in fluorescence that occurs due to protein turnover post-labeling rather than due to cellular proliferation.
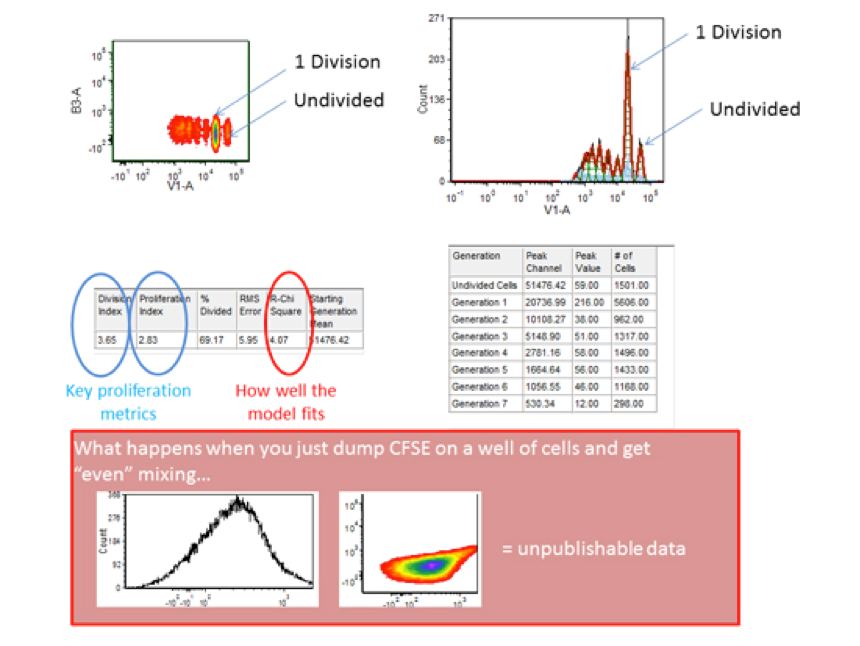
The above recommendations can prove the difference between extremely well resolved data and data that cannot be resolved into distinct generations of cellular proliferation. The ability to see “ideal” generations will be partly a function of the heterogeneity of the cells themselves (lymphocytes are depicted here).
Software programs like FlowJo and FCS Express are available to determine which cells are in which “generation” (i.e. generation 1 would be 1 round of mitosis). Ensure that the data is well fit (low RCS/reduced chi square value, etc), properly gated for single viable cells and then you can pay attention to various quantitative analytical parameters. The nomenclature changes with software packages but there are indexes to see how many rounds of division a cell has undergone on average if we either include or exclude non-dividing cells (limiting the assessment to either “responding” cells or all cells, respectively) as well as measurements of how many cells a dividing cell became, etc. For help with these analytical parameters and interpretation(s) please see the flow cytometry core staff.